Compute skinny scagnostic measure
sc_skinny(x, y)
# S3 method for default
sc_skinny(x, y)
# S3 method for scree
sc_skinny(x, y = NULL)
# S3 method for list
sc_skinny(x, y = NULL)Arguments
- x
numeric vector of x values
- y
numeric vector of y values
Value
A "numeric" object that gives the plot's skinny score.
Examples
require(ggplot2)
require(dplyr)
ggplot(features, aes(x=x, y=y)) +
geom_point() +
facet_wrap(~feature, ncol = 5, scales = "free")
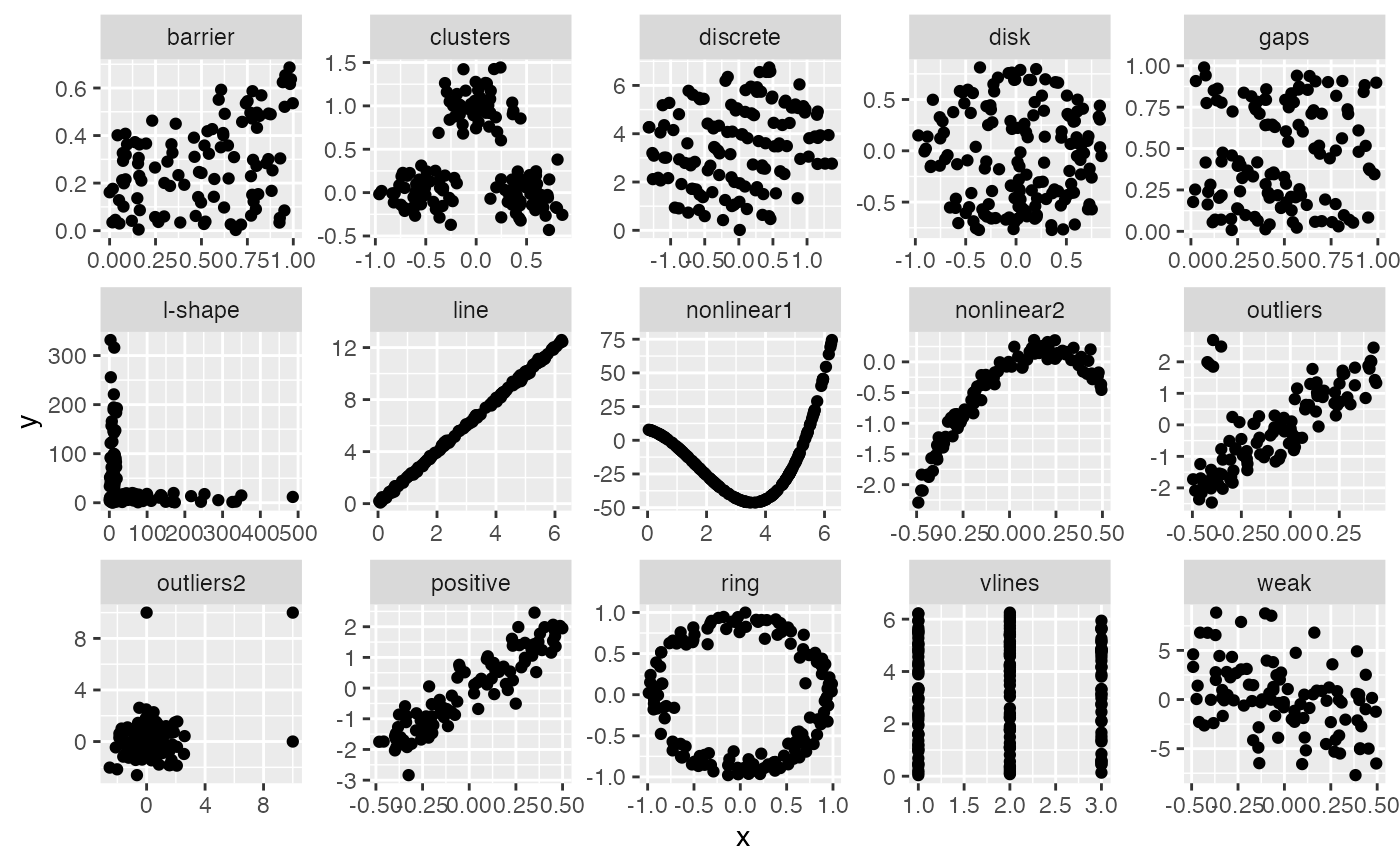 features %>% group_by(feature) %>% summarise(skinny = sc_skinny(x,y))
#> # A tibble: 15 × 2
#> feature skinny
#> <chr> <dbl>
#> 1 barrier 0.242
#> 2 clusters 0.536
#> 3 discrete 0.0711
#> 4 disk 0.104
#> 5 gaps 0.219
#> 6 l-shape 0.727
#> 7 line 0.861
#> 8 nonlinear1 0.307
#> 9 nonlinear2 0.672
#> 10 outliers 0.466
#> 11 outliers2 0.246
#> 12 positive 0.393
#> 13 ring 0.709
#> 14 vlines 0.127
#> 15 weak 0.148
sc_skinny(datasaurus_dozen_wide$away_x, datasaurus_dozen_wide$away_y)
#> [1] 0.2764928
features %>% group_by(feature) %>% summarise(skinny = sc_skinny(x,y))
#> # A tibble: 15 × 2
#> feature skinny
#> <chr> <dbl>
#> 1 barrier 0.242
#> 2 clusters 0.536
#> 3 discrete 0.0711
#> 4 disk 0.104
#> 5 gaps 0.219
#> 6 l-shape 0.727
#> 7 line 0.861
#> 8 nonlinear1 0.307
#> 9 nonlinear2 0.672
#> 10 outliers 0.466
#> 11 outliers2 0.246
#> 12 positive 0.393
#> 13 ring 0.709
#> 14 vlines 0.127
#> 15 weak 0.148
sc_skinny(datasaurus_dozen_wide$away_x, datasaurus_dozen_wide$away_y)
#> [1] 0.2764928